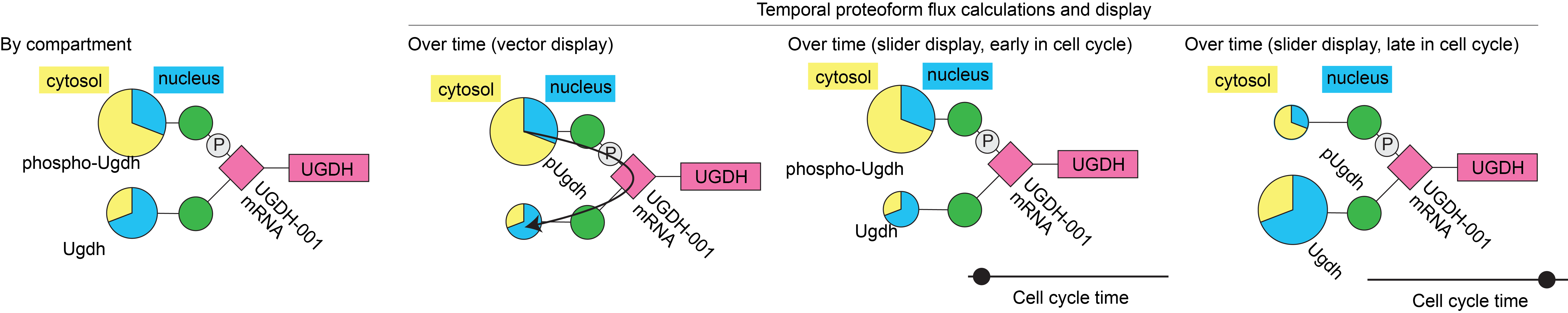
Area 2: Building a framework for quantifying proteoform family dynamics.
Proteoform families are defined by dynamic modifications with important functional consequences; this proteoform flux is thus worth investigating. However, tools for quantifying and visualizing the temporal flux between proteoforms do not yet exist. I will develop open-source tools to quantify and visualize the results of proteoform analysis. These new tools will allow the nascent field of proteoform systems biology to better illustrate important proteoformlevel differences. We will also use computer modeling of protein folding within proteoform families to better understand how proteoform variation impacts our ability to treat disease. Characterizing varied protein druggability will be an important step towards understanding variations in the success of disease treatments.